DNA stores all of the information necessary for life phenomena, and a cell transmits its own genetic information to two daughter cells through DNA replication and cell division. Replication stress can be caused by extracellular and intracellular sources during DNA replication, which leads to slowed or stalled replication. If cells do not properly cope with such risks, chromosome break and rearrangement will occur, resulting in genomic instability. That helps explain why replication stress is one of the major contributors to cancer development.
Although many DNA repair proteins function to protect and restart stalled replication process under stress condition, it is still unclear how replisome proteins, which are real players in DNA replication, contribute and communicate with those proteins to ensure faithful DNA replication. Led by Director Kyungjae Myung and Dr. Kyoo-young Lee, a research group at the Center for Genomic Integrity within the Institute for Basic Science (IBS) at UNIST revealed that ATAD5 actively deals with replication stress, in addition to its known function to prevent such stressful situations. Though ATAD5 has been known as a tumor suppressor by maintaining genomic stability and suppressing tumorigenesis, it has been unclear whether the replication regulatory protein is also involved in the replication stress response. “We have identified the fundamental mechanism of replication stress control, which is a major cause of cancer. Hopefully our work will contribute to the development of cancer therapy,” explains Director Myung.
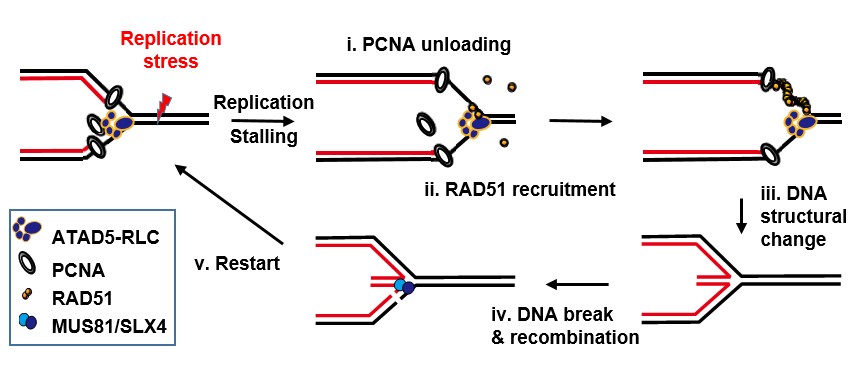
▲ Figure 1. In the face of DNA reapplication stress, ATAD5 i) removes PCNA from the stressed DNA and simultaneously ii) recruits RAD51 recombinase through protein-protein interaction. Then iii) RAD51 wraps and transforms the DNA into a stable structure. Subsequently, iv) DNA is cleaved via MUS81/SLX4 and recombined, and v) DNA replication restarts. This process keeps DNA stable under replication stress and ensures entire replication is complete.
Director Myung’s group recently found that ATAD5 regulates the functions of PCNA, one of main replication proteins by removing the ring-shaped PCNA from DNA to ensure a complete end of its cycling. ATAD5-depleted cells show characteristic features of replication stress such as a slow replication rate. Although the PCNA-regulating activity of ATAD5 may be the cause of this cellular response to its depletion, the researchers hypothesized that ATAD5 may have an additional role in counteracting replication stress. ATAD5 proteins play an important role in the DNA replication process itself, so experimentally reducing the amount of ATAD5 in cells results in many abnormalities associated with normal DNA replication. Because replication stress, which is focused in this study, also occurs during the DNA replication, it is critical to separate the effects of ATAD5 deficiency on the replication stress and the general DNA replication process. To overcome this, the researchers devised an experimental method to induce ATAD5 deficiency at the beginning of replication stress.
Under these experimental conditions, they found that, when ATAD5 level is reduced, cells cannot resume DNA replication stalled by replication stress, but increases genome instability such as intracellular chromosome breakage and micronucleated reticulocytes in mouse blood. This means that ATAD5 contributes to maintaining genomic stability by restoring DNA replication under replication stress. Thereafter, they conducted experiments to elucidate the molecular mechanism for ATAD5 restarting DNA replication. RAD51 plays critical roles in structural changes and stability of stalled replication sites under replication stress. The researchers found that ATAD5 promotes RAD51 recruitment to stalled replication sites by direct protein-protein interaction.
In addition, they reported that PCNA unloading by ATAD5 is a prerequisite for efficient RAD51 recruitment. These suggest that a series of processes starting with RAD51 recruitment and leading to structural changes, breakage, and eventual replication restart are regulated by ATAD5. These findings highlight that the role of ATAD5 in maintaining genome stability extends beyond its roles in PCNA unloading during normal DNA replication. Dr. Su Hyung Park, the first author of this study notes, “Mutations in the ATAD5 gene are frequently found in many cancer cells. This study contributes to understating the cause of tumor formation by ATAD5 mutations.”
Replication stress can be caused by extracellular sources such as oncogenes and chemicals. However, recent studies have shown that intracellular sources such as DNA-specific structures or the abnormal function of specific proteins can also provoke replication stress. In addition to enhancing our understanding of initial cellular responses to replication stress, this study provides us a novel conceptual advance that a protein can play dual roles in managing replication stress: the preemptive prevention and the resolution of replication stress. Dr. Kyoo-young Lee, the corresponding author of this study says, “In the future, we will examine how the internal factors cause replication stress and how the cells selectively recognize and cope with different stressor.”
The findings of this research have been published in Nature Communications on December 16, 2019.
Kyungjae Myung
Distinguished Professor, School of Life Sciences, UNIST
IBS Center for Genomics Integrity
Dahee Carol Kim
Public Information Officer, IBS
T: +82-42-878-8133
E:clitie620@ibs.re.kr
Story Source
Materials provided by Institute of Basic Science.
Notes for Editors
The online version of the original article can be found HERE.
Journal Reference
Su Hyung Park, “ATAD5 promotes replication restart by regulating RAD51 and PCNA in response to replication stress,” Nat Commun 10, 5718 (2019) doi:10.1038/s41467-019-13667-4