A research team, affiliated with UNIST has developed a novel CRISPR-based genome labeling scheme for reliable visualization of genomic elements in living cells. This labeling scheme, according to the research team, would enable real-time visual tracking of changes in chromatin structure.
CRISPR, also known as CRISPR/Cas9, is a type of gene-editing technology has been used to correct specific disease-causing DNA mutations. It consists of two components, which includes the Cas9 enzyme that functions as a pair of molecular scissors and a guide RNA (gRNA or sgRNA), which guides the Cas9 to the planned part of the genome, ensuring the right place is cut. Genome imaging based on the CRISPR system has attracted strong interest in recent years, as a powerful approach, yet has limited applicability due to background signals and non-specific aggregation of fluorophores within nuclei.
To address this issue, Professor Hajin Kim and his research team in the Department of Biomedical Engineering (BME) at UNIST has developed a novel CRISPR-based genome labeling scheme for reliable visualization of genomic loci in live cells for an extended period of time. To achieve this, the research team integrated tripartite superfolder green fluorescent protein (sfGFP) and the SunTag system into the design of a CRISPR complex. According to the research team, this labeling scheme allowed them to detect genomic loci with a greatly reduced background level and high target specificity, thereby enabling successful tracking of genomic loci with a small number of sequence repeats in live cells. Thanks to this, the structural changes within chromatin domains can be observed in real-time.
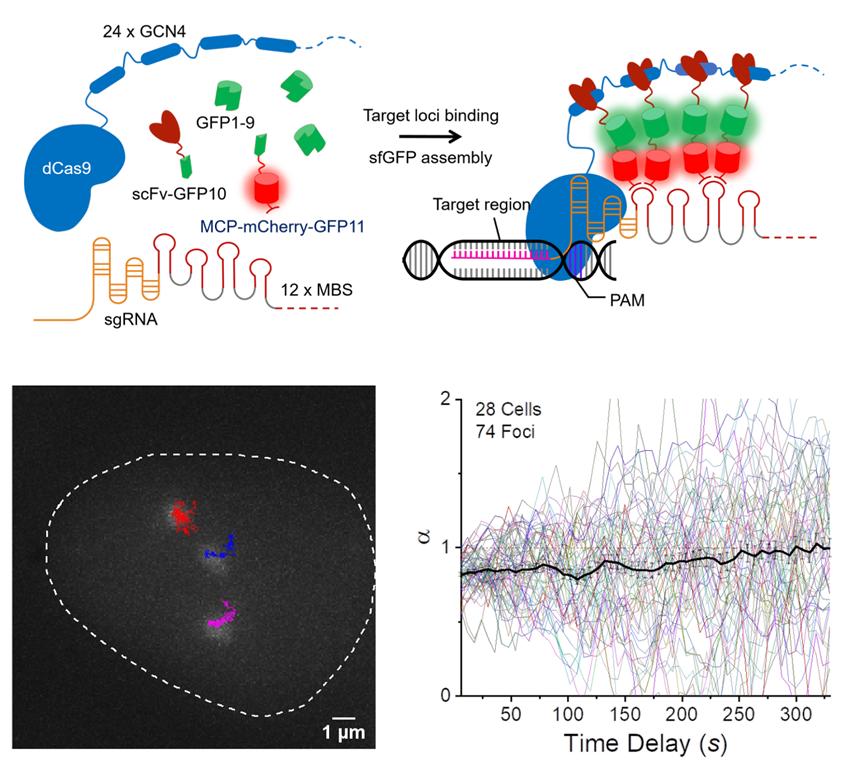
Figure 1. SunTag split-sfGFP CRISPR system enables background suppression and signal amplification for genome imaging. (A) Schematic design of the CRISPR-dCas9 system, integrated with tripartite sfGFP and the SunTag. (B) Representative trajectories of three C9-1 foci in a single nucleus, shown along with the firstframe image of GFP channel, traced from a 2D-projected z-stack movie of live AD-293 cells integrated with the SunTag split-sfGFP CRISPR system. The trajectories represent the loci movement for 30 min with 6-sec frame intervals. (C) Collected trajectories of C9-1 foci (n = 74).
Chromatin is a complex of genomic DNA and proteins that form chromosomes within the nucleus of eukaryotic cells. Each cell in the human body contains approximately 2 meters of DNA, yet the nucleus of a human cell, which contains the DNA, has diameters ranging from 6 to 10 μm. In recent years, there has been a growing interest in the study of chromatin structure and dynamics. That is because several studies have shown that aging is associated with changes in gene expression and chromatin structure, and that in many cases, including cancer.
“To identify the exact linkage among changes in chromatin structure, gene expression, aging, and cancer, it is necessary to develop new methods to track the movements of chromatin in real-time,” says Professor Kim.
In their study, the research team presented a new visualization scheme, combining tripartite fluorescent proteins with the SunTag system and demonstrated that it strongly suppressed background fluorescence and amplified locus-specific signals, allowing long-term tracking of genomic loci. Besides, with fluorescence analysis, the location and shape of DNA can also be determined. They also integrated the multicomponent CRISPR system into stable cell lines to allow quantitative and reliable analysis of dynamic behaviors of genomic loci. Due to the greatly elevated signal-to-background ratio, target loci with only small numbers of sequence repeats could be successfully tracked, even under a conventional fluorescence microscope. This feature enables the application of CRISPR-based imaging to loci throughout the genome and opens up new possibilities for the study of nuclear processes in living cells.
They also confirmed that DNA not only diffuses passively, as ink spreads in water, but also moves actively. This has been verified via long-term tracking of the movement of specific regions of DNA, using the newly-developed genome imaging technique. “Due to the thread-like structure of DNA, we expected it to show sub-diffusion when there is no active movement, but herein via long-term tracking of the C9-1 loci, we observed super-diffusion behavior,” says Narendra Chaudhary, the first author of the study.
This is also in line with recent research findings that DNA itself moves actively in the process of genetic information. Until now, it has been known that DNA does not act indirectly on other molecules. Rather, various enzymes act on DNA and copy its information into either more DNA, in DNA replication, or transcribe it into protein. Likewise, the enzymes in DNA repair pathways that act upon the damaged DNA substrate and allow for the genetic information stored in the DNA to be expressed in the physical and functional makeup of the cell.
The findings of this research have been published in the journal, Genome Research on September 4, 2020. This study has been supported by the National Research Foundation of Korea (NRF) and IBS, Korea.
Journal Reference
Narendra Chaudhary, Si-Hyeong Nho, Hayoon Cho, et al., “Background-suppressed live visualization of genomic loci with an improved CRISPR system based on a split fluorophore,” Genome Research, (2020).