Transcription factor is a protein that binds to a specific site of DNA and regulates the cell’s response so that genes can only be expressed when necessary. Creatures survive by efficiently responding to their surroundings using transcription factors with various characteristics. Recently, research results on the diversity of transcription factor control networks with such high importance have been published, drawing attention.
While global transcription factors (TFs) have been studied extensively in Escherichia coli model strains, conservation and diversity in TF regulation between strains is still unknown.
In search for the answer, Professor Donghyuk Kim and his research team in the School of Energy and Chemical Engineering at UNIST carried out a joint research with Professor Bernard Palsson from the University of California San Diego (UCSD). Based on ChIP-exo, one of the chromatin immune deposition experimental methods, and RNA-seq technology, the research team reconstructed regulon, a set of genes whose expression is regulated by the same transcription factor.
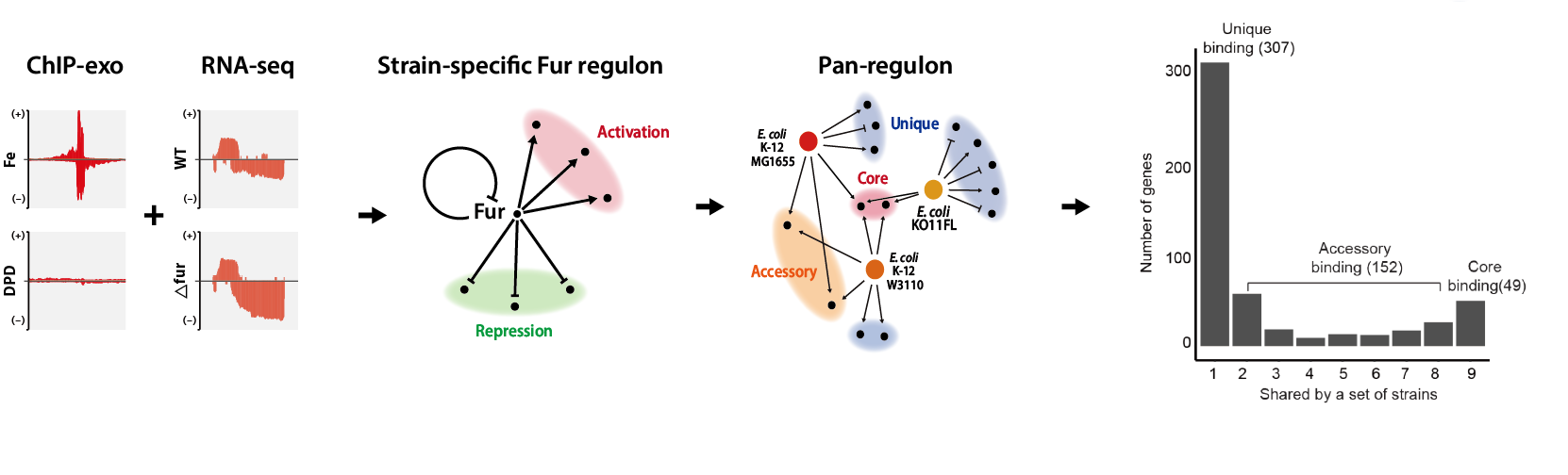
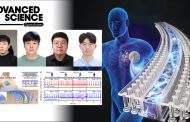
Figure 1. The set of commonly regulatory target genes of Fur is consistent and related to the regulation of iron metabolism. Meanwhile, the high number of uniquely regulatory targets of Fur showed diversity in Fur regulation among the nine E. coli strains. This diversity likely results in physiological differences between strains.
In particular, the research team conducted a study to analyze the preservation and regulatory diversity of iron absorption control transcription factors (Ferr). In particular, the transcription control network by Fur of nine strains that can encompass the characteristics of the entire E. coli was compared and analyzed. As a result, the research team created a new concept called “Pan-regulon,” which encompasses all 469 genes whose expression can be controlled by Fur in nine strains.
Through the analysis method, the research team divided pan-regulantrons into core regulators (36 genes commonly found in all strains), accessory regulators (158 genes found in some strains), and intrinsic regulators (275 genes found only in a single strain). In other words, it was confirmed that there are common genes controlled by Fur in all nine strains through fan-regulanton, but there are also quite a few genes that appear only in certain strains.
This study reveals the functional characteristics of Fur’s common gene-regulated target along with the concept of fan-regulanton, which was first defined. The diversity of transcriptional regulation present between E. coli strains results in physiological differences between each strain. In particular, through this study, meaningful results were derived that could further enhance the understanding of the transcription factor, called ‘Fur.’
The findings of this study have been published in the April 2023 issue of Nucleic Acids Research. This study has been supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (MSIT).
Journal Reference
Ye Gao, Ina Bang, Yara Seif, et al., “The Escherichia coli Fur pan-regulon has few conserved but many unique regulatory targets,” Nucleic Acids Res., (2023).