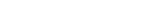For a protein to function appropriately, it must achieve its proper conformation and location within the crowded environment inside the cell. Moreover, information of subcellular locations of proteins is important for in-depth studies of cell biology, as well as understanding disease mechanisms and for developing novel drugs. Therefore, it is crucial to determine the subcellular localization of a given protein of interest (POI) in order to correctly understand their functions.
A research team, led by UNIST Professor Hyun-Woo Rhee of Natural Sciences has successfully developed a simple, robust, and efficient method for predicting the subcellular localization of proteins.
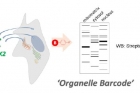
Through the study, Prof. Rhee and his team demonstrated ascorbate peroxidase (APEX)-generated molecular pattern recognition to provide information regarding intracellular microenvironments in living cells.
Using APEX labeling of specific proteins, the team has succeeded in identifying the subcellular localization of a protein of interest (POI) at the sub-compartment level, which has been considered challenging with traditional optical or biochemical methods.
Song-Yi Lee, a combined master’s-doctoral researcher in Prof. Rhee’s lab and the first author of the study states, “With the conventional biochemical methods and even recently developed super-resolution imaging methods, it has been difficult to provide sufficient resolution to discriminate whether target proteins or target sites are located on the cytosolic or luminal side.”
She adds, “Using this method, we successfully identified the membrane topology of HMOX1 on the ER membrane and sub-mitochondrial localization of MGST and TRMT61B, as well as several other mitochondrial proteins.”
They expect that this new method can be expanded to confirm sub-mitochondrial localization and membrane topologies of previously identified mitochondrial proteins.
According to the team, “Our method could be useful to the biology research community because it is easy to implement and provides an efficient solution to determining membrane topology and sub-organelle protein localization.”
This work has been supported by the UNIST research fund and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare of Korea. The results of the study have been appeared in the May 24th issue of Cell Reports, the leading journal of cell biology and the highest-impact journal in the world.
Journal Reference
Song-Yi Lee, Myeong-Gyun Kang, Jong-Seok Park, Geunsik Lee, Alice Y. Ting, and Hyun-Woo Rhee. “APEX Fingerprinting Reveals the Subcellular Localization of Proteins of Interest”, Cell Reports, (2016). 8.