“The amount of data we generate everyday is truly mind-boggling. In other words, depending on which analytical strategy you establish, you can make important discoveries and thus gain better understanding of cell conditions or diseases.”
A research team, led by Professor Dougu Nam in the School of Life Sciences at UNISThas presented a novel approach to identify cancer suppressing microRNA (miRNA) targets and relevant cellular signaling pathways through large-scale gene-expression data analysis.
MicroRNAs (miRNAs) are a class of short, non-coding RNA molecules (containing about 19-23 nucleotides) that post-transcriptionally control gene expression. Emerging studies have shown that miRNAs play a crucial role in many biological processes, thus providing beneficial diagnostic and prognostic information for patients with chronic diseases—including cancer and diabetes.

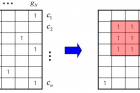
Biclustering equation, which allows clustering of rows and columns of a dataset simultaneously.
Through this study, Professor Nam and his team have developed a new analytical strategy that utilizes publicly available gene expression datasets, collected over a period of 15 years. To this end, the team has harnessed over 5,000 human microarray expression datasets based on a variety of cell conditions, such as disease, chemical treatment, tissues, and differentiations. They, then, analyzed the information on target genes of interest according to miRNA sequences. As a result, they could construct the so-called BiMIR, a bicluster database that provides the condition-specific microRNA target predictions. In total, 29,898 biclusters for 459 human miRNAs were collected in the BiMIR database where biclusters are searchable for miRNAs, tissues, diseases, keywords and target genes. (Click HERE to be redirected to the website of BiMIR).
“Applying this biclustering approach to large-scaled gene expression data allows precise identification of miRNA-directed regulatory networks that occur in various cell conditions, such as stem cells or certain types of diseases,” says Professor Nam. “In other words, it becomes possible to predict which genes are associated with the expression of breast cancer and what miRNA inhibit those genes.”
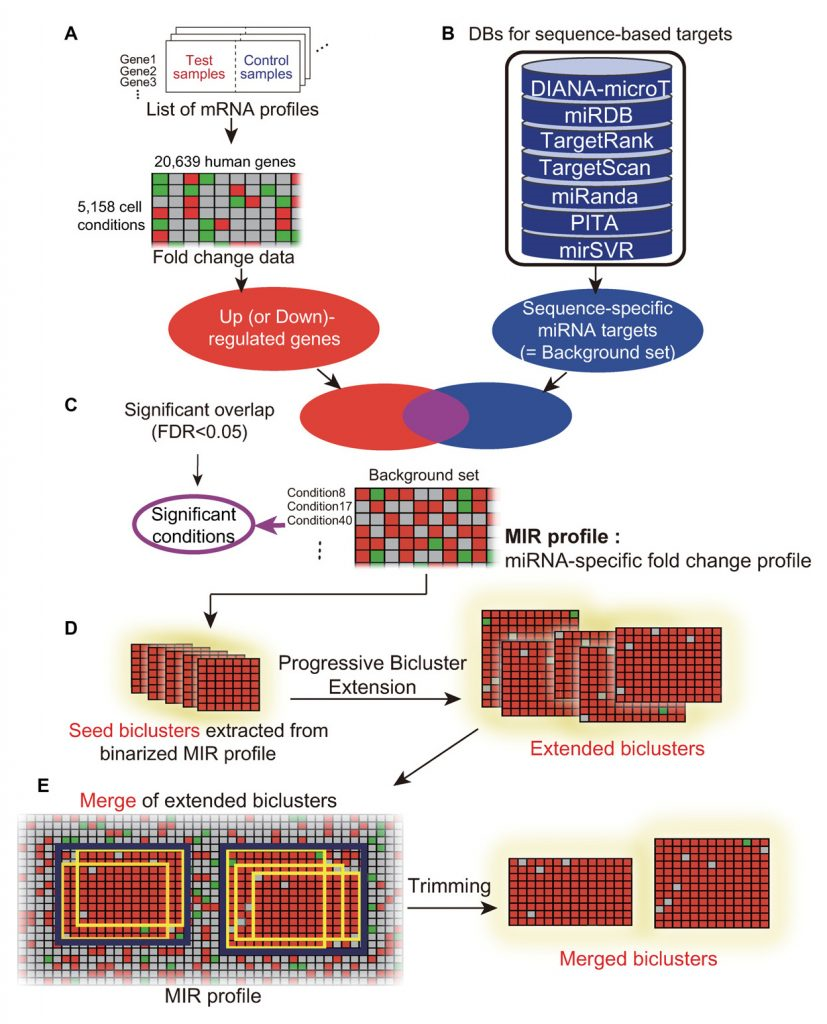
Above is theschematic image that shows the overview of the biclustering-based miRNA target prediction.
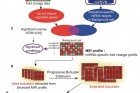
Through experiments, the research team analyzed cancer-specific biclusters and found that PI3K/Akt signaling pathway was intensively targeted by a few miRNAs in two cancer types (breast cancer and diffuse large B-cell lymphoma). Further, prognostic values of those miRNAs and the regulatory effects of miR-29 were also validated by Professor Nam in collaboration with Professor Jiyoung Park in School of Life Sciences at UNIST. Those results demonstrate that biclustering analysis is able to reveal key pathways controlled by miRNAs in disease.
“In breast cancer, bicluster targets of miR-29 were downregulated, thus did not regulate the PI3K/Akt signaling pathway,” says Professor Nam. “It was also experimentally validated that increasing the expression of miR-29 significantly reduced the target genes of interest, as well as the activation of the corresponding pathway.”
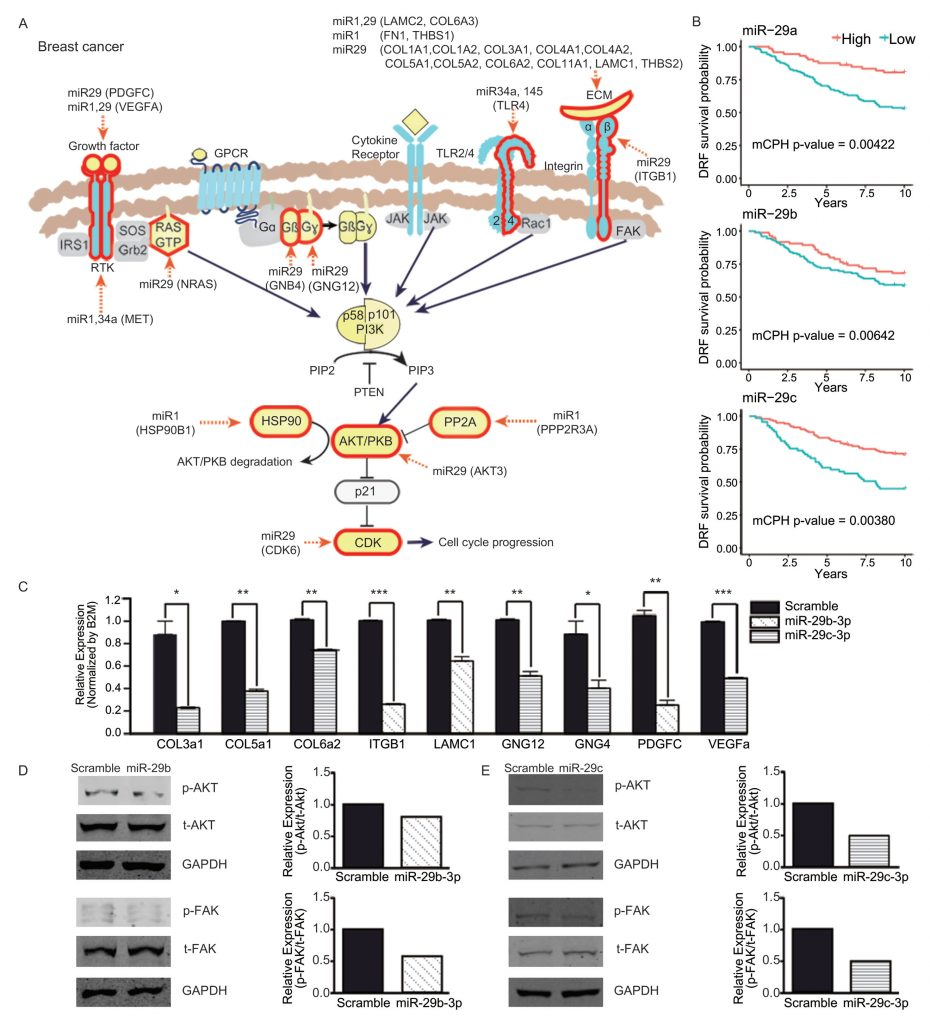
miRNA targets in PI3K/Akt signaling pathway (breast cancer).
“The biclustering approach presented here can also be applied for predicting the condition-specific targets of other sequence-specific regulators such as transcription factors or RNA-binding proteins,” says Professor Nam. “Our approach provides more reliable miRNA target groups that are robustly regulated across different cell conditions without using miRNA–mRNA profiles.”
Dr. Sora Yoon in the School of Life Sciences at UNIST also partook in the study, as the first author. This study has been supported by the Korea Post-Genome Project of the Ministry of Science and ICT (MSIT). Their findings have been published online in the March 2019 issue of Nucleic Acids Research (IF: 11.56), a prominent biology journal published twice monthly by Oxford University Press, Oxford, UK.
Journal Reference
Sora Yoon et al., “Biclustering analysis of transcriptome big data identifies condition-specific microRNA targets,” Nuclieic Acids Research, (2019).