A new metabolic pathway has been identified for methanotrophs, a group of bacteria that grow on methane (CH₄, also called greenhouse gas), as a sole source of carbon and energy. It is expected to be the cornerstone of technology that transforms methane into useful resources, using microorganisms.
Professor Donghyuk Kim in the School of Energy and Chemical Engineering at UNIST, in collaboration with Professor Eun Yeol Lee in the Department of Chemical Engineering at Kyung Hee University, has unveiled the metabolic pathways of M. alcaliphilum 20Z grown on methane and methanol. This hydrocarbon-eating microbe is a promising platform strain for bioconversion of one-carbon (C1) substrates into value-added products.
“Methane, which contributes to global warming, is released in large quantities in industrial waste gas, dairy and shale gas extraction processes,” says Professor Kim. “Therefore, in order to efficiently convert methane into fuels and high value-added chemicals, having an accurate understanding of the metabolic process of methane-consuming bacteria is pivotal.”
Understanding the metabolic network of methanotrophs can not only contribute to the genetic manipulation, but also help regulate its metabolic pathways. This allows using methane or methanol as raw materials, required for the production of desired materials. Until now, it has been known that the metabolism of methane and methanol starts with the phenomenon, known as the direct oxidation of methane to methanol. However, this is not enough to understand the metabolic process of each substance.
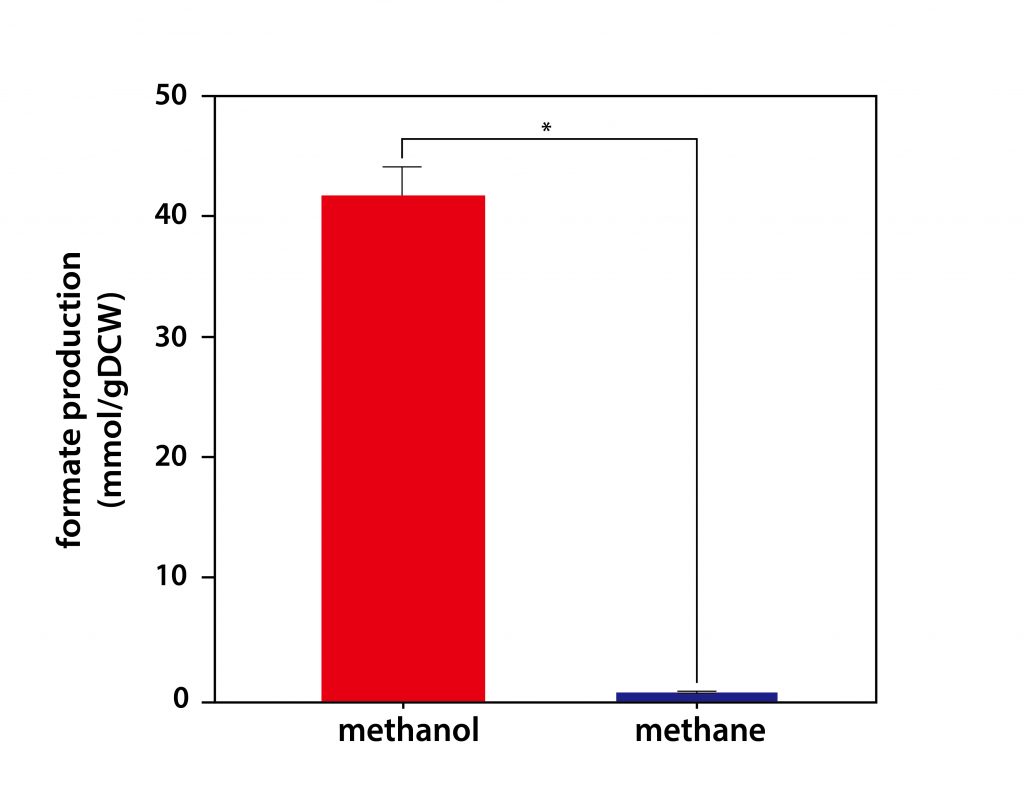
Figure 1. The accumulation of formate on methane and methanol grown cultures (mmol/gDCW). Data represent the means from 3 replicates ± standard deviations.
In this study, a genome-scale integrated approach was used to provide in-depth knowledge on the metabolic pathways of M. alcaliphilum 20Z grown on methane and methanol.
The research team found that when methanol bacteria use methanol as their carbon source, they produce more formic acid than they do with methane. The result is different even though the starting process is the same because the entire energy metabolic process changes. Because the amount of coenzyme (NADH) used in the chemical reaction to convert methane to methanol varies, there is a difference in total energy metabolism and carbon metabolism, and the resulting compound is different.
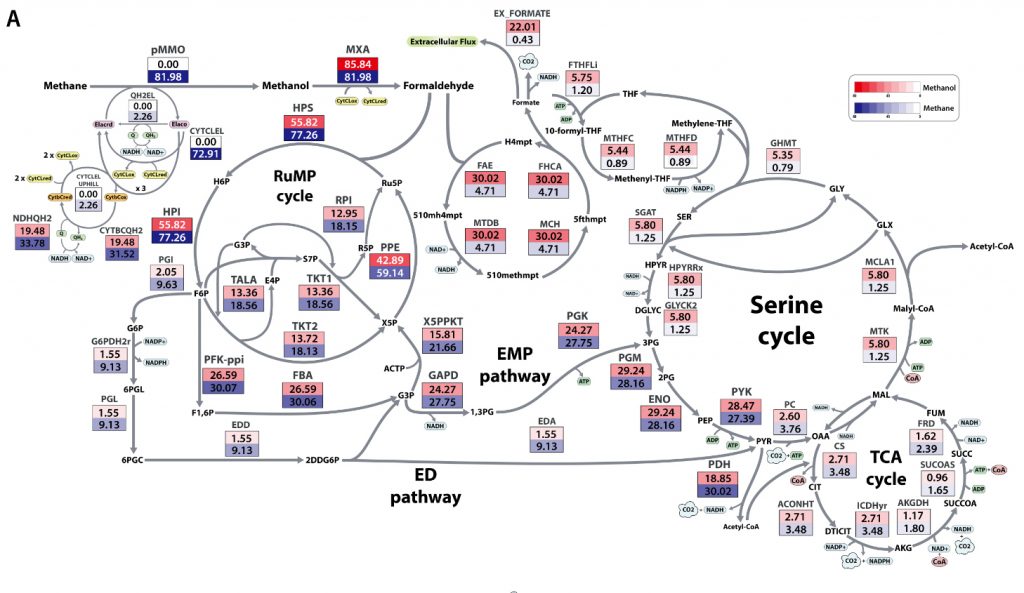
Figure 2. Normalized flux distribution of M. alcaliphilum 20Z on methane and methanol by Markov chain Monte Carlo sampling with the iIA407 model
The research team used a genome-scale integrated approach to provide in-depth knowledge on the metabolic pathways of M. alcaliphilum 20Z grown on methane and methanol. The metabolic process of M. alcaliphilum 20Z is as complex as playing ‘Ghost Leg’ games with many outcomes. Yet, thanks to genome-scale metabolic model (GSM), it became possible to see the core carbon metabolism pathways of M. alcaliphilum 20Z growth on methane and methanol at a glance.
“These findings provided a systems-level understanding of carbon metabolism and new opportunities for strain design to produce relevant products from different C1-feedstocks,” says Professor Kim.
The findings of this study have been published in Metabolic Engineering on October 15, 2019. This study has been supported by the C1 Gas Refinery Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science and ICT (MSIT).
Journal Reference
Anh Duc Nguyen, Joon Young Park, In Yeub Hwang, et al., “Genome-scale evaluation of core one-carbon metabolism in gammaproteobacterial methanotrophs grown on methane and methanol,” Matabolic Engineering, (2019).