Numerous anti-cancer drugs function by targeting the DNA within cancerous cells, halting their proliferation. Yet, cancer cells occasionally develop mechanisms to repair the damage inflicted by these drugs, diminishing their effectiveness. Consequently, physicians are increasingly embracing a novel approach to cancer treatment, known as precision medicine. This method involves selecting medications that precisely align with the unique attributes of an individual’s cancer. Precision medicine proves particularly beneficial in addressing cancers that have evolved to evade conventional treatments.
Trabectedin, a promising drug derived from the sea squirt Ecteinascidia turbinata, has shown potential in combating cancers resistant to conventional treatments. However, its precise mechanism of action has remained elusive—until now. A collaborative effort, led by Dr. Kook Son and Professor Orlando D. Schärer from the Center for Genomic Integrity within the Institute for Basic Science (IBS) at UNIST in South Korea, along with Dr. Vakil Takhaveev and Professor Shana Sturla from ETH Zurich, Switzerland, has illuminated the inner workings of this mysterious compound.
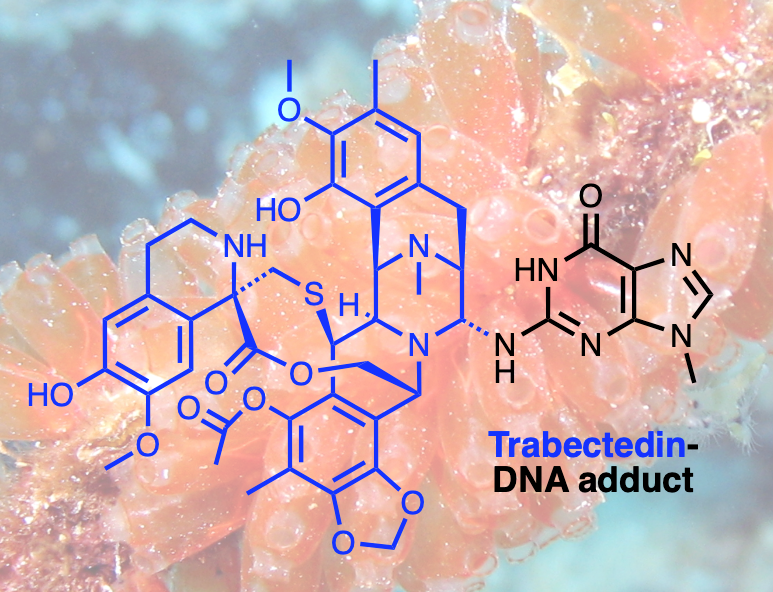
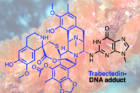
Figure 1. Trabectedin, an anti-cancer drug originally isolated from the sea squirt Ecteinascidia turbitana, is known to form cytotoxic DNA adducts. It is used to treat sarcomas and ovarian cancers and, in contrast to most antitumor agents, exerts its full activity in cells with active DNA repair.
Using highly sensitive high throughput COMET Chip assays to detect breaks formed in the genomes of cells, IBS researchers revealed trabectedin induces persistent breaks in the DNA of cancer cells. The researchers showed that these DNA breaks are only formed in cells with high levels of DNA repair, specifically those that operate a pathway called transcription-coupled nucleotide excision repair (TC-NER).
TC-NER is a vital mechanism that identifies DNA damage during transcription, initiating repair processes involving several endonucleases, like ERCC1-XPF and XPG. Trabectedin’s DNA damage disrupts this process by allowing the initial incision by ERCC1-XPF, but blocking the subsequent action by XPG, halting the TC-NER process. This disruption of the repair process leads to long-lasting DNA breaks that ultimately kill cancer cells.
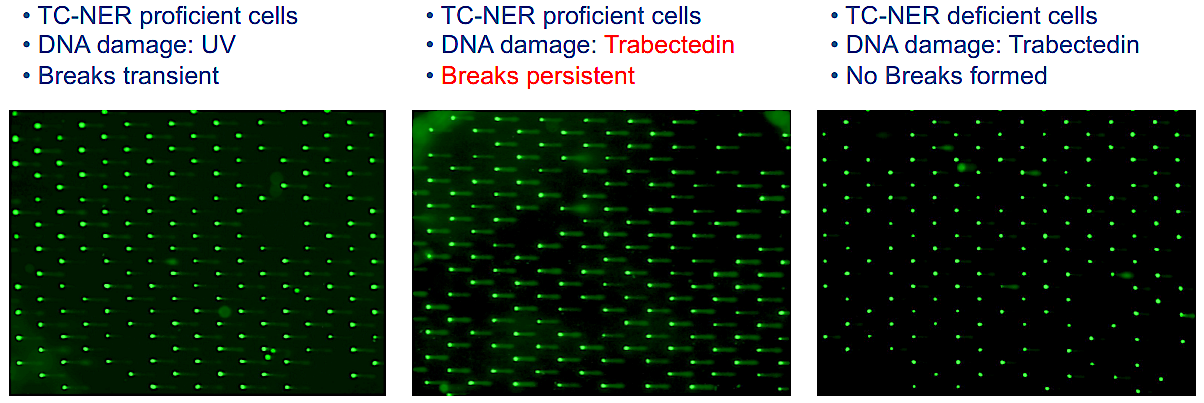
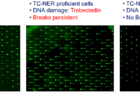
Figure 2. COMET Chip assays were used to measure Trabectedin-induced breaks in cells. Each green dot is a single nucleus of a cell and the tail length and the fraction of total DNA in the tail (comet) emerging from each nucleus is proportional to the number of breaks formed. Following UV treatment (left), few breaks are visible as UV lesions are excised from DNA by nucleotide excision repair (NER). Following treatment with trabectedin (center), DNA breaks persist due to an abortive NER reaction. The breaks are dependent on NER as they do not occur in TC-NER deficient cells (right) with inactivated XPF gene.
Analysis of the DNA break patterns induced by trabectedin revealed that breaks are formed throughout the genome, but only at sites where active transcription and with it, TC-NER occurs. Using this new insight into the mechanism of how DNA breaks are accumulated, the researchers sought to determine where in the genome these breaks occur. This led to the development of a new method called TRABI-Seq (for TRABectedin-Induced break sequencing), which allows for the precise identification of trabectedin’s action sites within tumor cell DNA.
“This incision by ERCC1-XPF creates a markable free hydroxyl group in the DNA, enabling us to sequence DNA and locate these breaks,” explains Dr. Son.
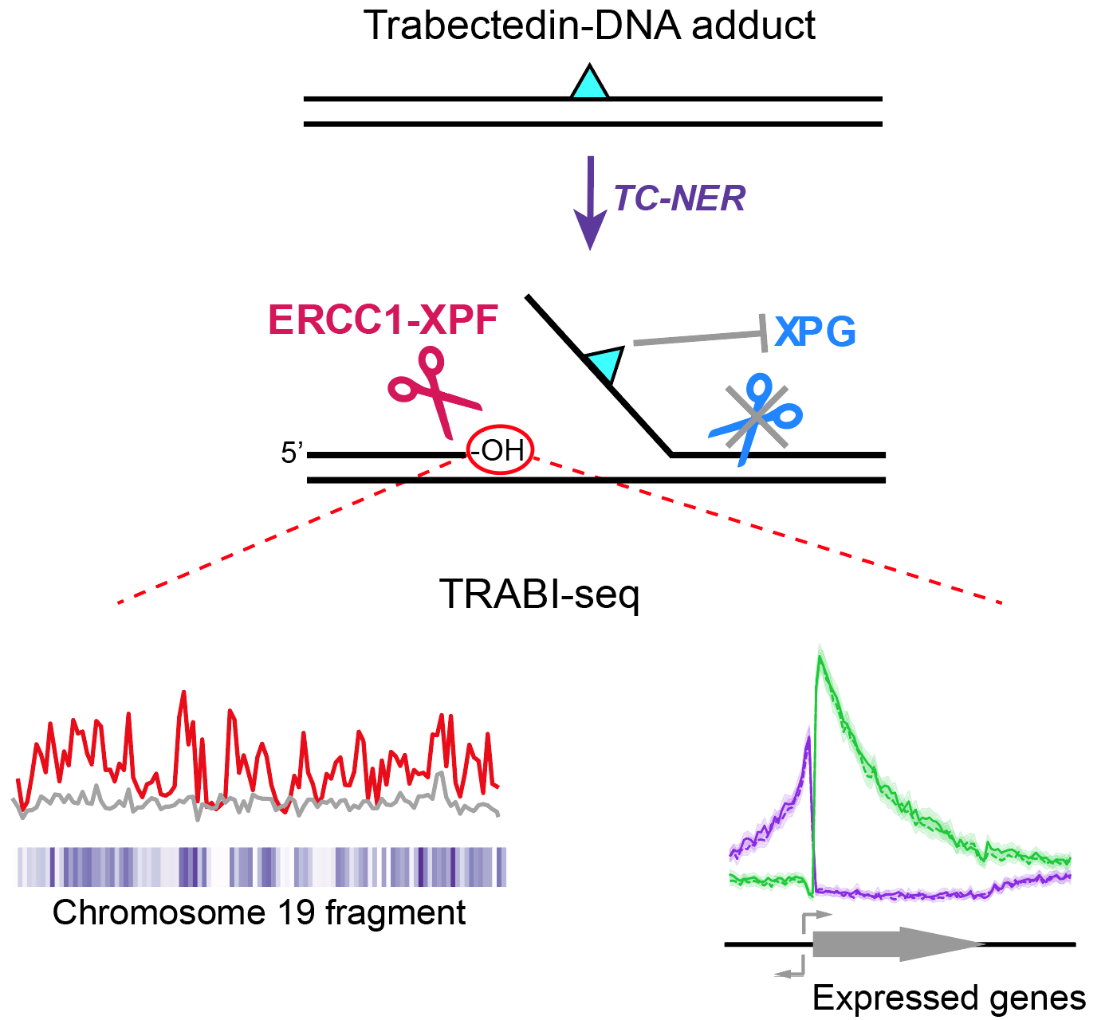
Figure 3. Summary of the mechanism of trabectedin-induced TC-NER-mediated break formation and break mapping by TRABI-Seq. Trabectedin-DNA adducts are recognized by TC-NER and lead to an abortive reaction as these adducts block the incision of the XPG endonuclease, causing persistent XPF-mediated breaks. TRABI-Seq (TRABectidin-Induced break sequencing) was used to map those breaks in a genome-wide fashion. The distribution of these persistent breaks was mostly ascribed to highly transcribed regions of the genome. This approach will now be used to determine how trabectedin induces DNA breaks in genomes of various cancer cell lines to develop TRABI-Seq as a diagnostic tool for cancer therapy.
TRABI-Seq is being tested on various cancer cells to determine trabectedin’s efficacy in targeting tumors with advanced DNA repair capabilities, often associated with elevated transcription levels due to oncogene activation. It is hoped that these findings will help position trabectedin as both a predictive marker to identify vulnerable cancers and a therapeutic option for precision treatment. With its ability to target tumors resistant to conventional therapies, trabectedin may provide further hope in the fight against drug-resistant cancer with advanced DNA repair capabilities.
Their findings have been published in the online version of Nature Communications (IF 16.6) on February 15, 2024.
Orlando D. Schärer
Associate Director, IBS Center for Genomics Integrity
E: orlando.scharer@ibs.re.kr
William Suh
IBS Communications Team
T: +82-42-878-8137
E: willisuh@ibs.re.kr
Story Source
Materials provided by Institute of Basic Science.
Notes for Editors
This press release is made available courtesy of the IBS Communications Team. The online version of the original article can be found HERE.
Journal Reference
Kook Son, Vakil Takhaveev, Visesato Mor, et al., “Trabectedin derails transcription-coupled nucleotide excision repair to induce DNA breaks in highly transcribed genes,” Nat. Commun., (2024).