Like the hurdle runners, the principle of protein movement is found by jumping around DNA. Using this principle of rapid movement and damage detection, it is expected that clues can be found in the treatment of various genetic diseases, including cancer.
A research team, led by Professor Jayil Lee in the School of Life Science at UNIST in collaboration with Distinguished Professor Orlando Schärer, has succeeded in observing the movement of the human NER protein, XPC-RAD23B. Their findings have been published in the journal, Nucleic Acids Research (IF: 11.56) on August 2, 2019.
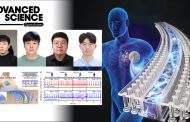
XPC-RAD23B protein is known to play a role in detecting DNA damage in our bodies. However, it has not been confirmed how the protein finds damage. The team used a single-molecule spectroscopy technique called “DNA curtain” to see the movement of the XPC-RAD23B protein on DNA in real time. The results showed that the protein moved along the DNA and identified the site of damage, and it was also observed that it jumped around to avoid other proteins on the DNA.
Model for lesion search mechanism of XPC-RAD23B.
“The XPC-RAD23B was able to easily pass through other proteins, so it was able to quickly find damage to up to 3 billion DNA sites,” says Na Young Cheon, the first author of the study. “This will enable us to develop molecular biological methods to quickly identify DNA damage that causes various diseases.”
DNA containing genetic information is easily damaged and modified by UV or toxic substances. These mutations don’t have mutations because the nucleotide ablation repair (NER), which quickly and accurately restores damaged DNA in our bodies, continues.
This repair is the interaction of various proteins, the starting point for the XPC-RAD23B protein is to identify the site of damage. The researchers hope the discovery will serve as a molecular biological basis for treating a variety of genetic diseases, including skin cancer and pigmentary dry skin caused by DNA damage.
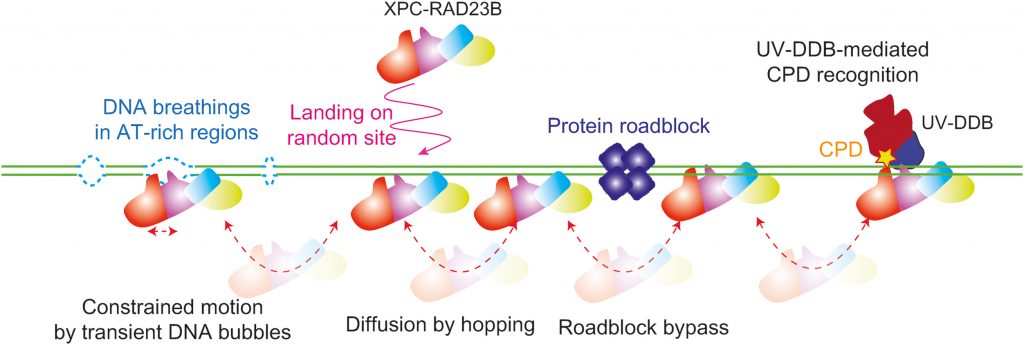
 ▲Schematic of DNA curtain experiment for the collision between XPC-RAD23B and EcoRI. EcoRIE111Q structure is adopted from protein data bank (PDB ID: 1CL8). Kymograph for the collision between XPC-RAD23B (red) and EcoRIE111Q (green). The green arrow represents EcoRI cognate site on λ-DNA.
▲Schematic of DNA curtain experiment for the collision between XPC-RAD23B and EcoRI. EcoRIE111Q structure is adopted from protein data bank (PDB ID: 1CL8). Kymograph for the collision between XPC-RAD23B (red) and EcoRIE111Q (green). The green arrow represents EcoRI cognate site on λ-DNA.
“Current microscopic techniques do not allow us to accurately observe the interaction of proteins and DNA in the cell nucleus,” says Professor Lee. “It provides the foundation for observing more precisely the interaction of DNA and protein that causes various diseases.”
This study has been supported by the Mid-career Research Program through the National Research Foundation (NRF), the POSCO Science Fellowship of POSCO TJ Park Foundation, and the Korean Institute for Basic Science (IBS). It has been also featured as NAR’s Breakthrough Articles.
NAR’s Breakthrough Articles present high-impact studies answering long-standing questions in the field of nucleic acids research and/or opening up new areas and mechanistic hypotheses for investigation. These articles are chosen by the Editors on the recommendation of Editorial Board Members and Referees. They represent the very best papers published at NAR.
Journal Reference
Na Young Cheon et al., “Single-molecule visualization reveals the damage search mechanism for the human NER protein XPC-RAD23B,” NAR, (2019).