In the pursuit of understanding the pathogenic expression mechanisms of bacteria and the advancements in biofoundry technology, identifying and analyzing protein-DNA binding sites is crucial. Researchers at UNIST and Korea University have developed a novel method known as ChIP-mini, which allows for precise identification of these binding sites using significantly fewer samples than previously possible.
Professor Donghyuk Kim from the School of Energy and Chemical Engineering at UNIST and Dr. Eun-Jin Lee from the Department of Life Sciences at Korea University have collaborated to develop an optimized ChIP-mini method—a low-input ChIP-exo that utilizes 5,000 times fewer bacterial cells than traditional approaches—enabling precise identification of binding sites for DNA-binding proteins in host-infected pathogens.
This innovative method, based on Chromatin Immunoprecipitation (ChIP)—a technique utilized to isolate DNA fragments bound by proteins—allows for high-resolution analysis, achieving base pair-level precision (approximately 0.34 nanometers) by examining as few as 4.8 million cells. Consequently, ChIP-mini can accurately identify individual binding sites, even when multiple proteins bind in close proximity, in stark contrast to the most recent conventional ChIP-exo experiments, which required between 10 billion and 100 billion cells to achieve similar precision.
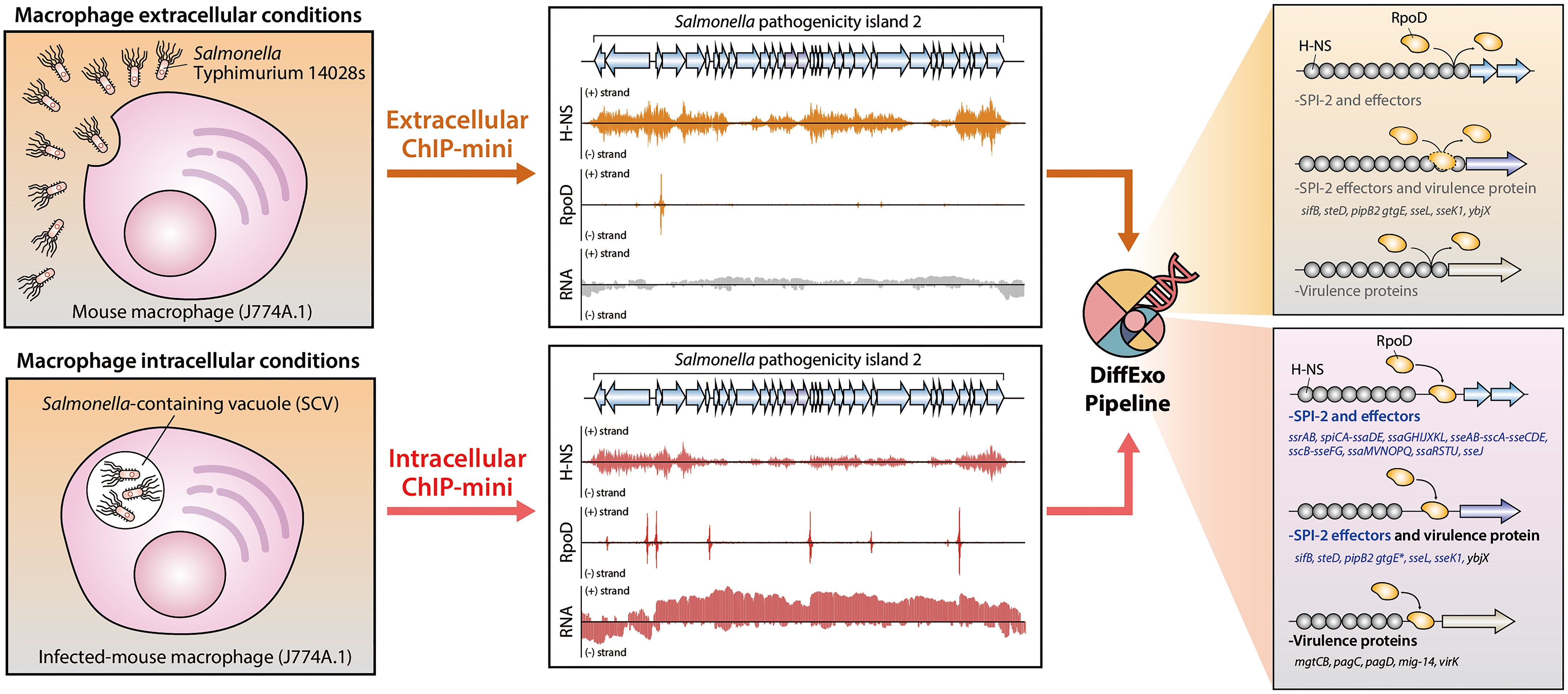
Figure 1. A schematic diagram with an overview of the study design and the main procedures from publication.
The research team validated ChIP-mini by isolating trace amounts of Salmonella bacteria, a known infectious agent, and quantitatively analyzing changes in DNA binding positions and intensities of two crucial proteins, H-NS and RpoD. Salmonella initially inhibits pathogenic gene expression by strongly binding H-NS to DNA outside host macrophages but reduces H-NS binding and activates pathogenic genes via RpoD (RNA polymerase sigma factor 70) upon entering the host cell, thereby avoiding detection by the immune system.
Previous ChIP experiments struggled with insufficient quantities of Salmonella within host cells due to its low abundance at the site of infection. To address this challenge, the research team employed DiffExo, a statistical program developed for quantitative analysis.
Additionally, the cost of individual analyses using ChIP-mini is approximately 20,000 KRW, constituting a twelvefold reduction compared to earlier methods. Dr. Jon Young Park, co-first author at UNIST, stated, “If integrated with Next Generation Sequencing (NGS) automation platforms, this technology could facilitate rapid and cost-effective generation of extensive datasets essential for biofoundry applications.” He further added, “We are actively pursuing research to ensure compatibility with NGS automation devices.”
Biofoundry technology involves producing high-value proteins by engineering microorganisms, similar to semiconductor manufacturing processes, and requires extensive datasets for effective microbial gene editing and optimal circuit design.
Professor Kim noted, “This research serves as a foundational technology in the biofoundry field, particularly in identifying gene expression networks of infectious microorganisms and discovering novel bio-parts.”
The findings of this research have been published in the Nucleic Acids Research on February 10, 2025. This study has been supported by the National Research Foundation of Korea (NRF) and the Ministry of Science and ICT (MSIT).
Journal Reference
Joon Young Park, Minchang Jang, Eunna Choi, et al., “ChIP-mini: a low-input ChIP-exo protocol for elucidating DNA-binding protein dynamics in intracellular pathogens,” Nucl. Acids Res., (2025).