A research team, affiliated with the Korea Genomics Center (KOGIC) at UNIST has released data from the initial phase of the Korean Genome Project (Korea1K), including information describing 1,094 whole genomes, along with 79 quantitative clinical traits. Their findings have been published in the journal, Science Advances on May 27, 2020.
Korea1K is the largest genome sequencing project, being carried out as part of the Genome Korea in Ulsan, which was launched in 2015. The project has recently released the first large-scale data to construct a genetic map and diversity analysis of Koreans. The researchers plan on securing genomic data of 10,000 individuals by 2020, which they said can be a useful resource for clinical and ethnogenetic studies.
In the first stage of the project, 1,094 whole genomes were sequenced at an average depth of 31x. The data for each sequenced genome has been paired with 79 associated traits from the person whose genome was sequenced. Besides, through the comparison of the human genome reference (hg38) that was released in 2003, the research team identified 39 million single-nucleotide variants (SNVs) and indels of which half were singleton or doubleton and detected Korean-specific patterns based on several types of genomic variations.
“In order to better explain the function and role of Korean-specific or rare and low-frequency variants in the genetic landscape of Koreans, it is imperative to secure large-scale Korean variome database,” says Director Semin Lee of Korean Genomics Center (KOGIC).
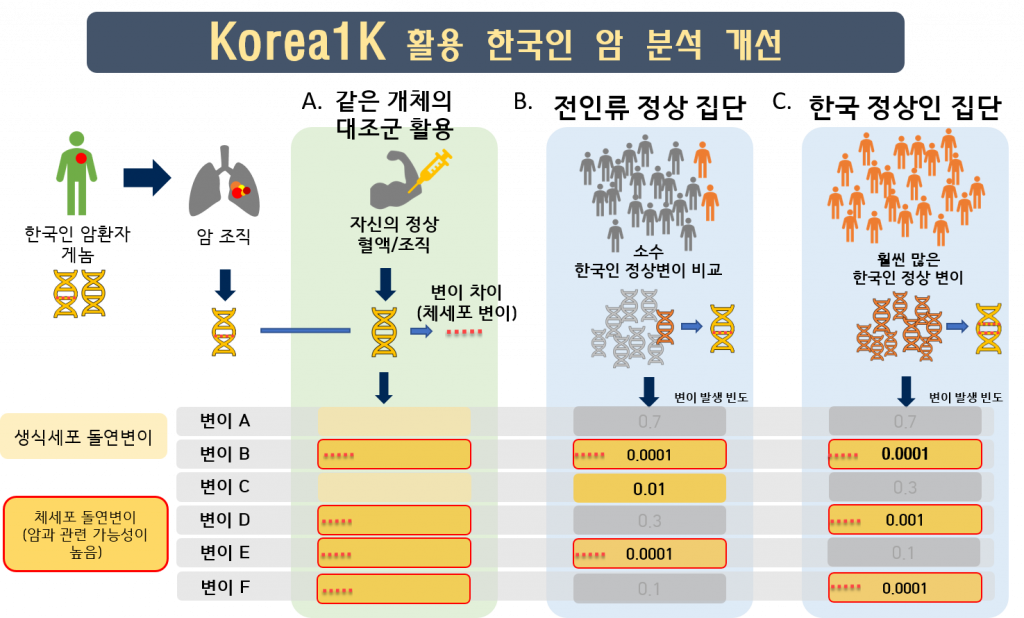
Throughout the study, researchers found that using Korea1K as a reference resulted in better imputation accuracy for Koreans than there had been with the 1000 Genomes Project (1KGP*) panel. In particular, the Korea1K set had the highest accuracy of prediction of cancer-associated genetic variants. Meanwhile, KGP is a joint project by the Personal Genome Project at Harvard Medical School, the National Center for Standard Reference Data of Korea, Clinomics Inc., and the Korean Genomics Center of UNIST. The research team noted, “As proof of utility, germline variants in cancer samples could be filtered out more effectively when the Korea1K variome was used as a panel of normals compared to non-Korean variome sets.”
“Overall, this study shows that Korea1K can be a useful genotypic and phenotypic resource for clinical and ethnogenetic studies,” says Yeongsong Choi (School of Life Sciences, UNIST) who performed the analysis.
The researchers noted that of the 1,094 Korean genomes in the dataset, 1,007 genomes were newly generated in combination with systematically acquired clinical and biochemical measurements from the blood and urine of the participants. The research team used these data to characterize single-nucleotide variants (SNVs), indels, copy number variations (CNVs), transposable element (TE) insertion, and human leukocyte antigen (HLA) type in the Korean population and contrasted the Korean data with similar data from other populations.
According to the research team, approximately half of the variants that they identified were classified as singleton or doubleton, with more than 70% of them had not been previously reported in dbSNP. On the other hand, fewer than 20% of the variants were designated as very common. They also found that, with regard to indels, the Korea1K set displayed more deletions than insertions, possibly resulting from skewed variant calling.
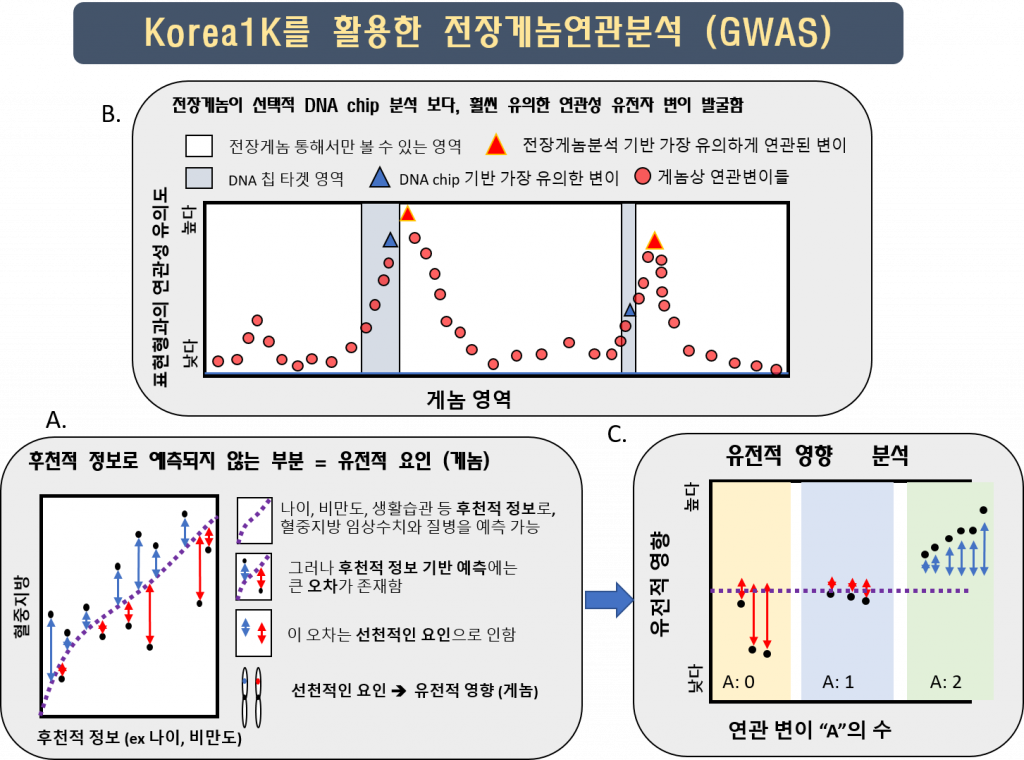
In the Korea1K set, researchers were also able to quantify 79 quantitative traits measured from 984 samples through health checks provided to the KGP participants, including the tests that measure the level of triglycerides and thyroid hormone levels in the blood. The analysis resulted in 467 variants that were statistically linked via genome-wide association study (GWAS) to 11 quantitative traits. The 467 variants were clumped into 15 independent loci on eight chromosomes, and 11 of them contained previously reported variants linked to a trait. Of the 15 independent loci, 4 were newly identified in this study and 9 contained variants with a higher correlation than previously known.
“While many of the previous GWAS studies focused on associating specific genetic variations with particular diseases, the large-scale Korean variome database constructed herein is potentially applicable in studies on various cancers and other diseases of Koreans and can indirectly help reduce the cost of certain genetic analyses,” noted Sungwon Jeon and Youngjune Bhak (School of Life Sciences, UNIST), the first authors of the study. “This kind of personal whole-genome dataset combined with common health check–derived clinical information is possibly a good exemplary path for an ethnicity-relevant reference panel for future personalized medical applications for Koreans.”
Meanwhile, the Genome Korea in Ulsan safely manages all individuals’ genomic information collected from participants in voluntary consent, using pseudonymization and anonymization procedures. In this study, 1,094 whole genomes were sequenced that require a minimum of petabyte (1 PB) of storage space. Among the results of Korea1K variation analysis study, including the frequencies of genetic variation among Koreans can be accessed by anyone via Korea1K webpage (http://1000genomes.kr/).
Journal Reference
Sungwon Jeon, Youngjune Bhak, Yeonsong Choi, et al., “Korean Genome Project: 1094 Korean personal genomes with clinical information,” Science Advances (2020). DOI: 10.1126/sciadv.aaz7835











Pingback: UNIST and Ulsan to Announce Successful Completion of 100,000 Ulsan Genomes Project! – UNIST BME()
Pingback: UNIST and Ulsan to Announce Successful Completion of 100,000 Ulsan Genomes Project! — dup – UNIST BME()
Pingback: Outstanding Faculty and Staff Awards Honoring 2020 Winners! – UNIST BME()