“Each human cell contains approximately 2 meters of DNA, yet that are too small to be visible to the unaided eye. Amazingly, all of that DNA is tightly packaged into a teeny little nucleus. There, we were able to observe the most basic separation principle of oil and water phases.”
A new study by Professor Hajin Kim in the School of Life Sciences at UNIST presents the notion of “DNA Phase Separation”, which suggests that the DNA within the nucleus may trigger phase separation, like oil in water. Unlike existing biological methods, Professor Kim explains how DNA works through a basic principle of physics.
According to this principle, DNA is contained in the cell nucleus in a condensed and folded form, which creates spatially separated regions within it. Here, the genetic information that the cell will use immediately is made available and easy, while other remaining information are kept in compressed archive files. Thus, two or more regions of DNA create a separate space, providing a basis for cells to use information efficiently.
In Physics, this phenomenon is called ‘Phase Separation’. This phenomenon can be induced due to the changes in temperature, pressure, and constituent elements of a mixture between two immiscible liquid, solid, or gaseous phases. Recently, however, interesting phase separation phenomena have been reported in living organisms, in which various molecules, like water, protein, and lipid are mixed. There are some prevailing hypotheses, concerning Tau proteins undergo a liquid-liquid phase separation to from droplets of essentially liqid protein to cause Alzheimer’s disease or that RNA foci form by phase separation of the repeat-containing RNA is associated with Huntington’s disease.
“Our study suggests for the first time that the phase separation phenomenon, which used to be considered the exclusive property of Physics, can be also triggered by heavy methylation of DNA,” says Professor Kim. “This study showed the possibility that DNA conformation could determine cellular activities such as gene expression and stem cell differentiation at the molecular level.”
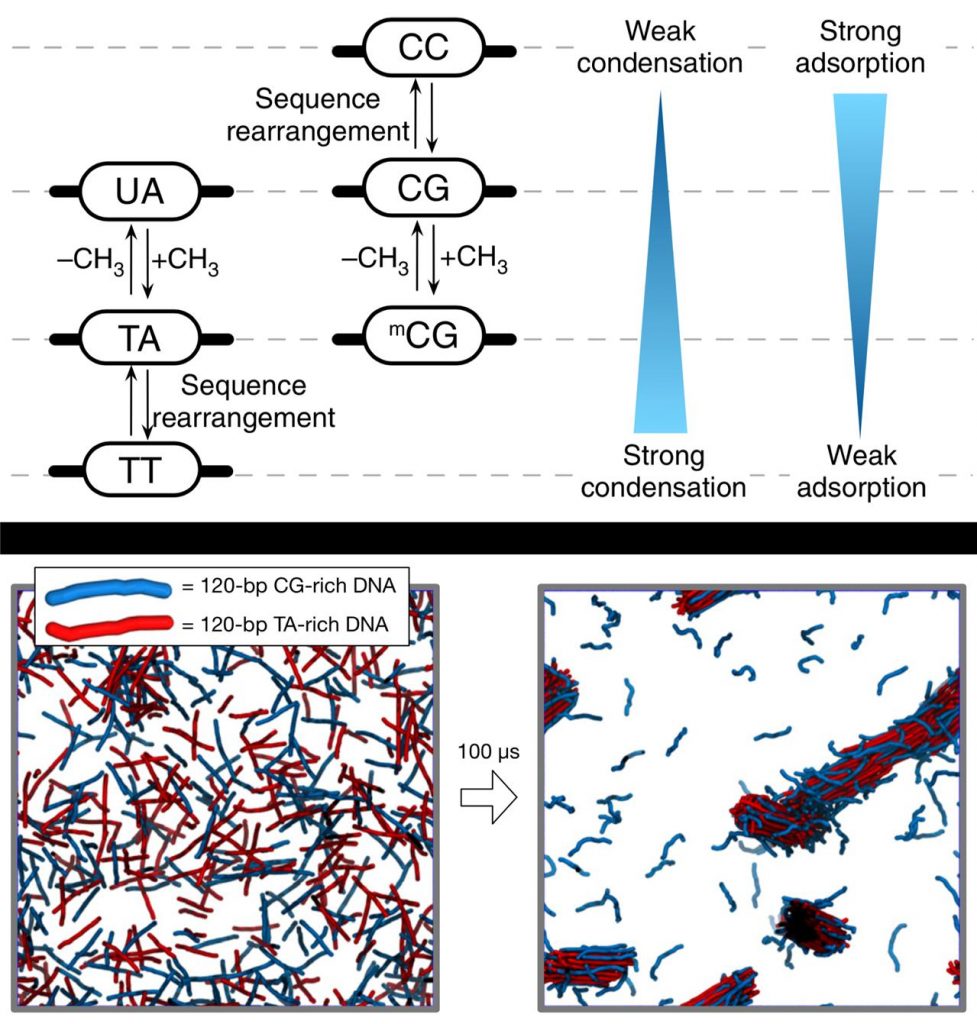
Above figure shows the phase separation of DNA in a mixture of dsDNA molecules driven by sequence-specific forces.
Electrostatic forces exist in DNA phase separation. Since DNA has a strong negative charge, there is a repulsive force between the negatively charged DNA. However, in the presence of a certain type of biological polycation, DNA molecules attract each other and such strength of inter-DNA attraction depends on DNA sequences. Such principle was once predicted and demonstrated by Professor Kim via simulation and published in the 2016 issue of Nature Communications. This time, the research team has experimentally proved that this principle is due to the interaction between polyamines and methyl groups of nucleic acid molecule.
DNA is composed of four different bases of nucleic acids—Adenine (A), Thymine (T), Cytosine (C), and Guanine (G), which are strung along ribbons of sugar- phosphate molecules in the shape of a double helix. These bases are always paried in such a way that Adenine connects to Thymine and Cytosine connects to Guanine. These pairings produce the following base pair possibilities: A-T, T-A, G-C, and C-G. Besides, through their MD simulations and experiments, the research team revealed that the DNA attraction strength was much stronger for the TA-repeat DNA pairs in comparison to the CG-repeat ones.
“Thymine contain C5 methyl groups and adding this to cytosine results in stronger attraction, almost as strong as the TA-rich DNA pairs,” says Professor Kim. “The C5 methyl groups attached to cytosine is a major chemical modification to DNA in epigenetics, and therefore we may expect the linkage between the epigenetic regulation of chromosome structure and the stem cell differentiation.”
The findings of this study has been published in Nucleic Acids Research (IF: 10.162), an internationally renowned molecular biology journal owned and published by Oxford University Press.
Journal Reference
Hyunju Kang, et al., “Sequence-dependent DNA condensation as a driving force of DNA phase separation,” Nucleic Acids Research, (2018).