Chromatin immunoprecipitation (ChIP) has been widely employed by researchers to determine the in vivo location of DNA binding sites on the genome for a particular protein of interest. In particular, ChIP-exo enables researchers to delineate genome-wide binding landscapes of DNA-binding proteins with near single base-pair resolution. However, the peak calling step hinders ChIP-exo application because the published algorithms tend to have higher false-positive and false-negative prediction rates.
To solve this, a research team, led by Professor Donghyuk Kim from the Department of Energy and Chemical Engineering at UNIST has introduced a novel machine learning-based ChIP-exo peak calling suite, also known as DEOCSU (DEep-learning Optimized ChIP-exo peak calling SUite).
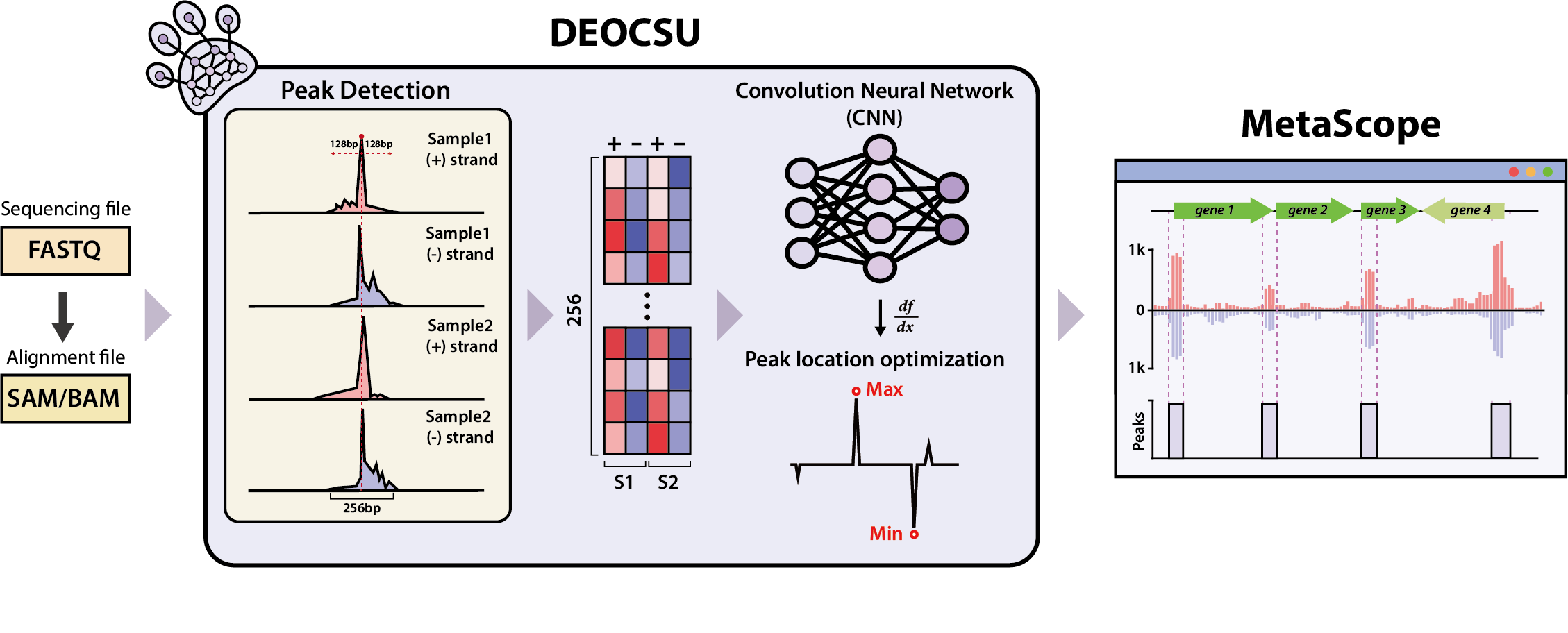
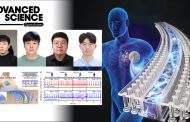
Figure 1. Overview of DEOCSU
The DEOCSU provides a highly accurate prediction of DNA binding sites from ChIP-exo datasets by distinguishing the visualized data of bona fide peaks from false ones using the deep CNN approach, according to the research team. The peak calling performance of DEOCSU was also compared with published tools, including MACE, MACE-elite, ChExMi, and PeakXus using the in-house RpoN ChIP-exo datasets of six E. coli strains and other bacteria (Klebsiella and Shigella). Performance validation of the trained deep-learning model indicated its high accuracy (96.2%), high precision (95.1%), and high recall (96.2%), said the research team.
Figure 2. Performance validation of DEOCSU using the ChIP-exo data of sigma factors in Escherichia coli K-12 str. MG1655.
According to the research team, applying the new suite to both in-house and publicly available ChIP-exo datasets obtained from bacteria, eukaryotes and archaea revealed an accurate prediction of peaks containing canonical motifs, highlighting the versatility and efficiency of DEOCSU.
“DEOCSU can be executed on a cloud computing platform or the local environment,” said the research team. “With visualization software included in the suite, adjustable options such as the threshold of peak probability, and iterable updating of the pre-trained model, DEOCSU can be optimized for users’ specific needs.”
The findings of this research have been published in the January 2023 issue of Briefings in Bioinformatics. This study has been supported by the National Research Foundation of Korea (NRF), funded by the Ministry of Science and ICT (MSIT). It has also been supported through the UNIST Center for Waste Plastics Carbon Cycling (UWCC), funded by the Circle Foundation.
Journal Reference
Ina Bang, Sang-Mok Lee, Seojoung Park, et al., “Deep-learning optimized DEOCSU suite provides an iterable pipeline for accurate ChIP-exo peak calling,” Brief. Bioinform., (2023).